The functions below are useful for labeling features/genes in plots.
Users have to call on aes(label = ...) or (label = ...) to define label's text
Based on the function, the label will be placed at a specific location:
geom_..._text()will plot text in the middle of the feature.geom_..._tag()will plot text on top of the feature, with a 45 degree angle.geom_..._note()will plot text under the feature at the left side.
The ... can be either replaced with feat or gene depending on which
track the user wants to label.
With arguments such as hjust, vjust, angle, and nudge_y, the user
can also manually change the position of the text.
Usage
geom_feat_text(
mapping = NULL,
data = feats(),
stat = "identity",
position = "identity",
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
geom_feat_tag(
mapping = NULL,
data = feats(),
stat = "identity",
position = "identity",
hjust = 0,
vjust = 0,
angle = 45,
nudge_y = 0.03,
xjust = 0.5,
strandwise = TRUE,
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
geom_feat_note(
mapping = NULL,
data = feats(),
stat = "identity",
position = "identity",
hjust = 0,
vjust = 1,
nudge_y = -0.03,
xjust = 0,
strandwise = FALSE,
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
geom_gene_text(
mapping = NULL,
data = genes(),
stat = "identity",
position = "identity",
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
geom_gene_tag(
mapping = NULL,
data = genes(),
stat = "identity",
position = "identity",
hjust = 0,
vjust = 0,
angle = 45,
nudge_y = 0.03,
xjust = 0.5,
strandwise = TRUE,
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
geom_gene_note(
mapping = NULL,
data = genes(),
stat = "identity",
position = "identity",
hjust = 0,
vjust = 1,
nudge_y = -0.03,
xjust = 0,
strandwise = FALSE,
...,
parse = FALSE,
check_overlap = FALSE,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)Arguments
- mapping
Set of aesthetic mappings created by
aes(). If specified andinherit.aes = TRUE(the default), it is combined with the default mapping at the top level of the plot. You must supplymappingif there is no plot mapping.- data
The data to be displayed in this layer. There are three options:
If
NULL, the default, the data is inherited from the plot data as specified in the call toggplot().A
data.frame, or other object, will override the plot data. All objects will be fortified to produce a data frame. Seefortify()for which variables will be created.A
functionwill be called with a single argument, the plot data. The return value must be adata.frame, and will be used as the layer data. Afunctioncan be created from aformula(e.g.~ head(.x, 10)).- stat
The statistical transformation to use on the data for this layer. When using a
geom_*()function to construct a layer, thestatargument can be used to override the default coupling between geoms and stats. Thestatargument accepts the following:A
Statggproto subclass, for exampleStatCount.A string naming the stat. To give the stat as a string, strip the function name of the
stat_prefix. For example, to usestat_count(), give the stat as"count".For more information and other ways to specify the stat, see the layer stat documentation.
- position
A position adjustment to use on the data for this layer. This can be used in various ways, including to prevent overplotting and improving the display. The
positionargument accepts the following:The result of calling a position function, such as
position_jitter(). This method allows for passing extra arguments to the position.A string naming the position adjustment. To give the position as a string, strip the function name of the
position_prefix. For example, to useposition_jitter(), give the position as"jitter".For more information and other ways to specify the position, see the layer position documentation.
- ...
Other arguments passed on to
layer()'sparamsargument. These arguments broadly fall into one of 4 categories below. Notably, further arguments to thepositionargument, or aesthetics that are required can not be passed through.... Unknown arguments that are not part of the 4 categories below are ignored.Static aesthetics that are not mapped to a scale, but are at a fixed value and apply to the layer as a whole. For example,
colour = "red"orlinewidth = 3. The geom's documentation has an Aesthetics section that lists the available options. The 'required' aesthetics cannot be passed on to theparams. Please note that while passing unmapped aesthetics as vectors is technically possible, the order and required length is not guaranteed to be parallel to the input data.When constructing a layer using a
stat_*()function, the...argument can be used to pass on parameters to thegeompart of the layer. An example of this isstat_density(geom = "area", outline.type = "both"). The geom's documentation lists which parameters it can accept.Inversely, when constructing a layer using a
geom_*()function, the...argument can be used to pass on parameters to thestatpart of the layer. An example of this isgeom_area(stat = "density", adjust = 0.5). The stat's documentation lists which parameters it can accept.The
key_glyphargument oflayer()may also be passed on through.... This can be one of the functions described as key glyphs, to change the display of the layer in the legend.
- parse
If
TRUE, the labels will be parsed into expressions and displayed as described in?plotmath.- check_overlap
If
TRUE, text that overlaps previous text in the same layer will not be plotted.check_overlaphappens at draw time and in the order of the data. Therefore data should be arranged by the label column before callinggeom_text(). Note that this argument is not supported bygeom_label().- na.rm
If
FALSE, the default, missing values are removed with a warning. IfTRUE, missing values are silently removed.- show.legend
logical. Should this layer be included in the legends?
NA, the default, includes if any aesthetics are mapped.FALSEnever includes, andTRUEalways includes. It can also be a named logical vector to finely select the aesthetics to display. To include legend keys for all levels, even when no data exists, useTRUE. IfNA, all levels are shown in legend, but unobserved levels are omitted.- inherit.aes
If
FALSE, overrides the default aesthetics, rather than combining with them. This is most useful for helper functions that define both data and aesthetics and shouldn't inherit behaviour from the default plot specification, e.g.annotation_borders().- hjust
Moves the text horizontally
- vjust
Moves the text vertically
- angle
Defines the angle in which the text will be placed. *Note
- nudge_y
Moves the text vertically an entire contig/sequence. (e.g.
nudge_y = 1places the text to the contig above)- xjust
Move text in x direction
- strandwise
plotting of feature tags
Value
A ggplot2 layer with gene text.
A ggplot2 layer with feature tags.
A ggplot2 layer with feature notes.
A ggplot2 layer with gene text.
A ggplot2 layer with gene tags.
A ggplot2 layer with gene notes.
Details
These labeling functions use ggplot2::geom_text() under the hood.
Any changes to the aesthetics of the text can be performed in a ggplot2 manner.
Examples
# example data
genes <- tibble::tibble(
seq_id = c("A", "A", "A", "B", "B", "C"),
start = c(20, 40, 80, 30, 10, 60),
end = c(30, 70, 85, 40, 15, 90),
feat_id = c("A1", "A2", "A3", "B1", "B2", "C1"),
type = c("CDS", "CDS", "CDS", "CDS", "CDS", "CDS"),
name = c("geneA", "geneB", "geneC", "geneA", "geneC", "geneB")
)
seqs <- tibble::tibble(
seq_id = c("A", "B", "C"),
start = c(0, 0, 0),
end = c(100, 100, 100),
length = c(100, 100, 100)
)
# basic plot creation
plot <- gggenomes(seqs = seqs, genes = genes) +
geom_bin_label() +
geom_gene()
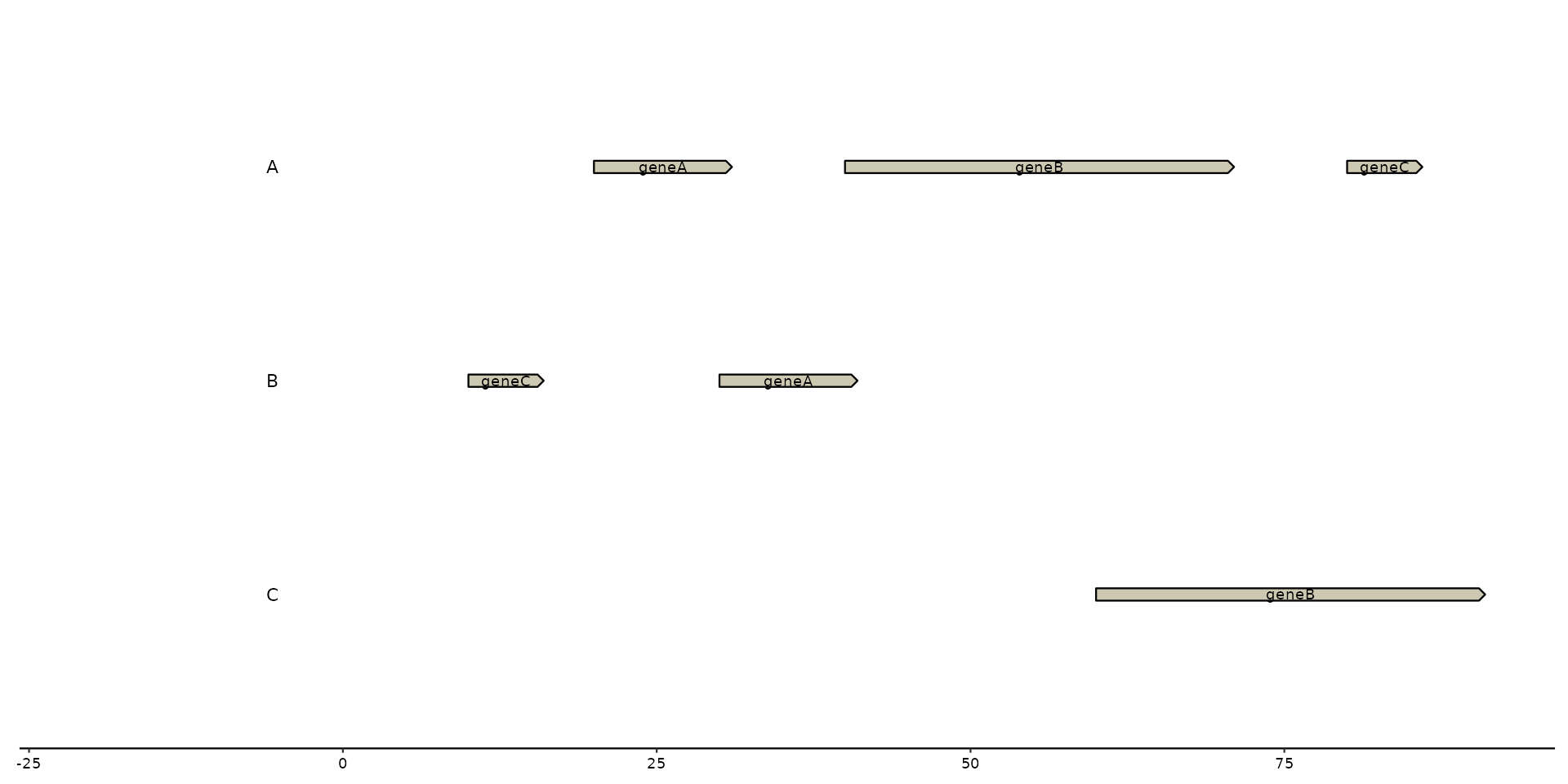
# geom_..._text
plot + geom_gene_text(aes(label = name))
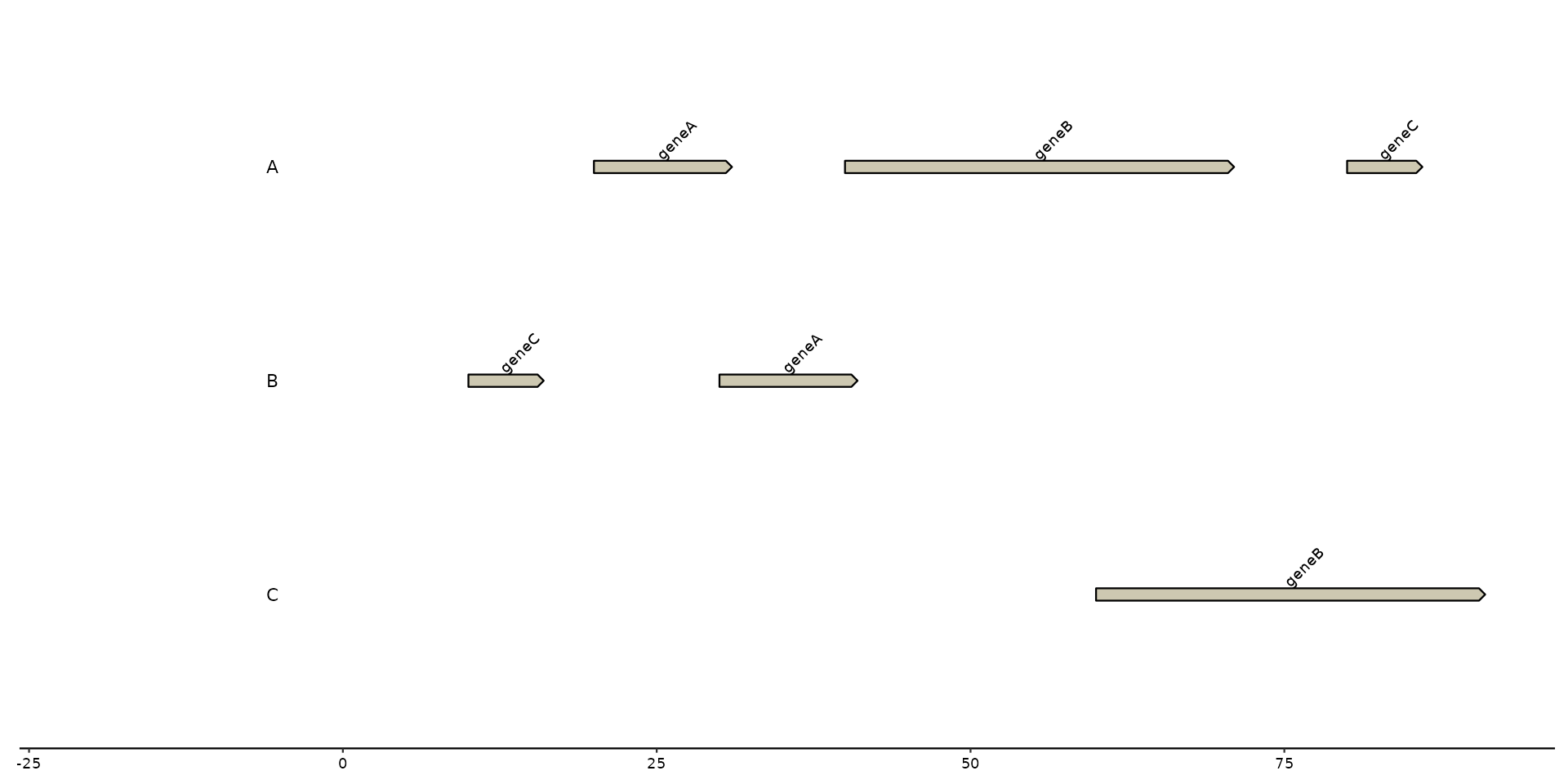
 # geom_..._tag
plot + geom_gene_tag(aes(label = name))
# geom_..._tag
plot + geom_gene_tag(aes(label = name))
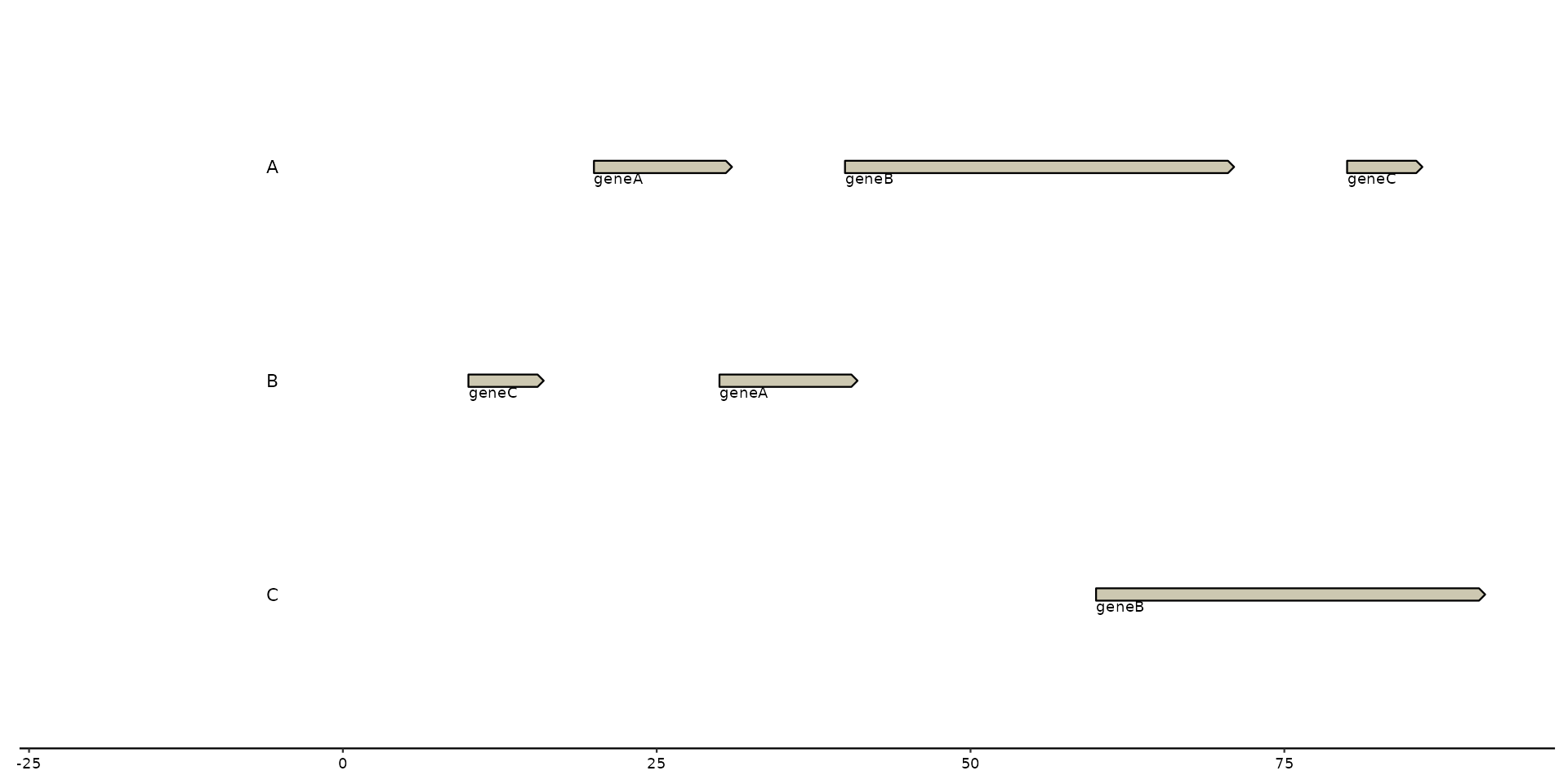
 # geom_..._note
plot + geom_gene_note(aes(label = name))
# geom_..._note
plot + geom_gene_note(aes(label = name))
 # with horizontal adjustment (`hjust`), vertical adjustment (`vjust`)
plot + geom_gene_text(aes(label = name), vjust = -2, hjust = 1)
# with horizontal adjustment (`hjust`), vertical adjustment (`vjust`)
plot + geom_gene_text(aes(label = name), vjust = -2, hjust = 1)
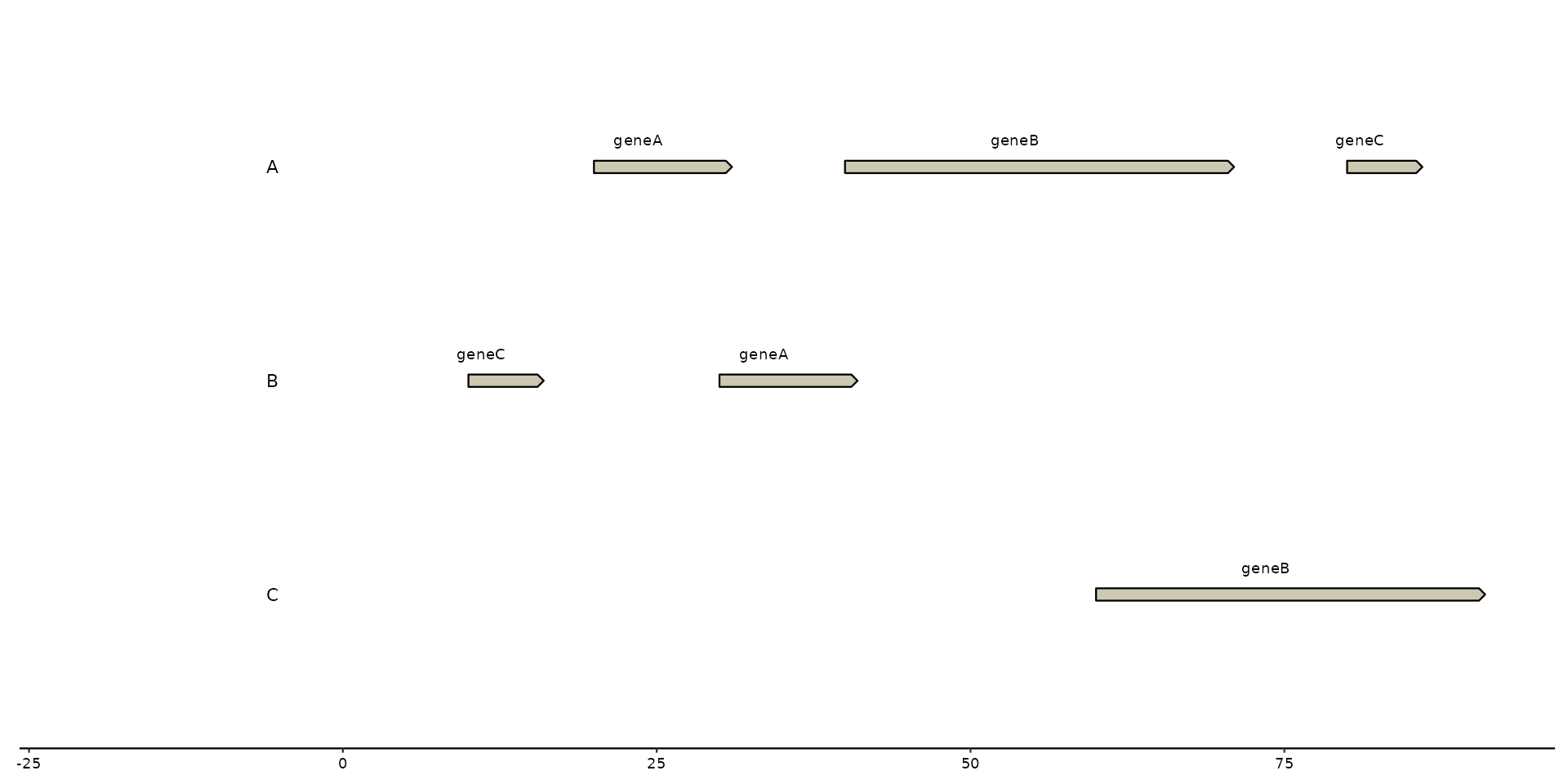 # using `nudge_y` and and `angle` adjustment
plot + geom_gene_text(aes(label = name), nudge_y = 1, angle = 10)
# using `nudge_y` and and `angle` adjustment
plot + geom_gene_text(aes(label = name), nudge_y = 1, angle = 10)
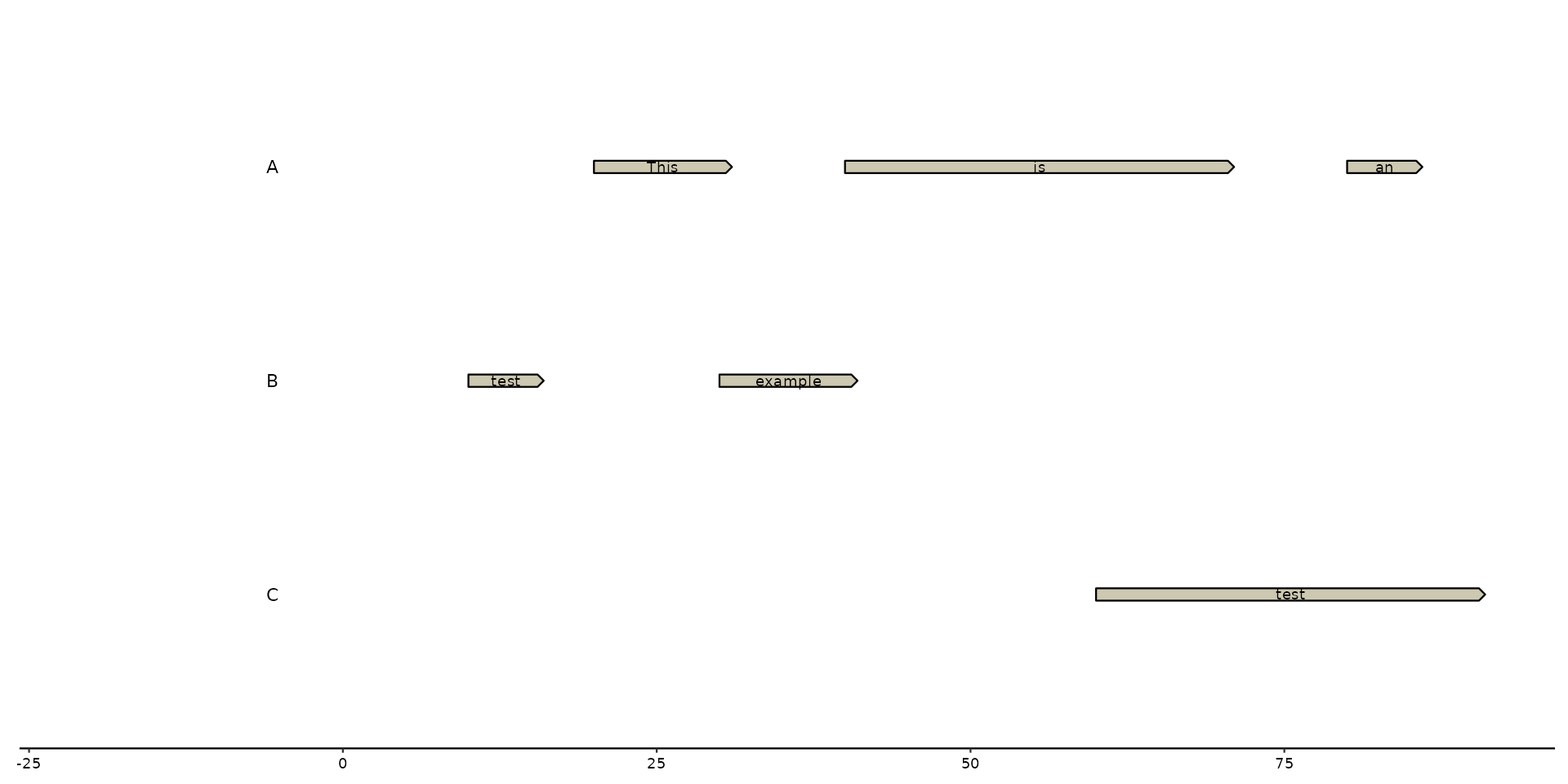
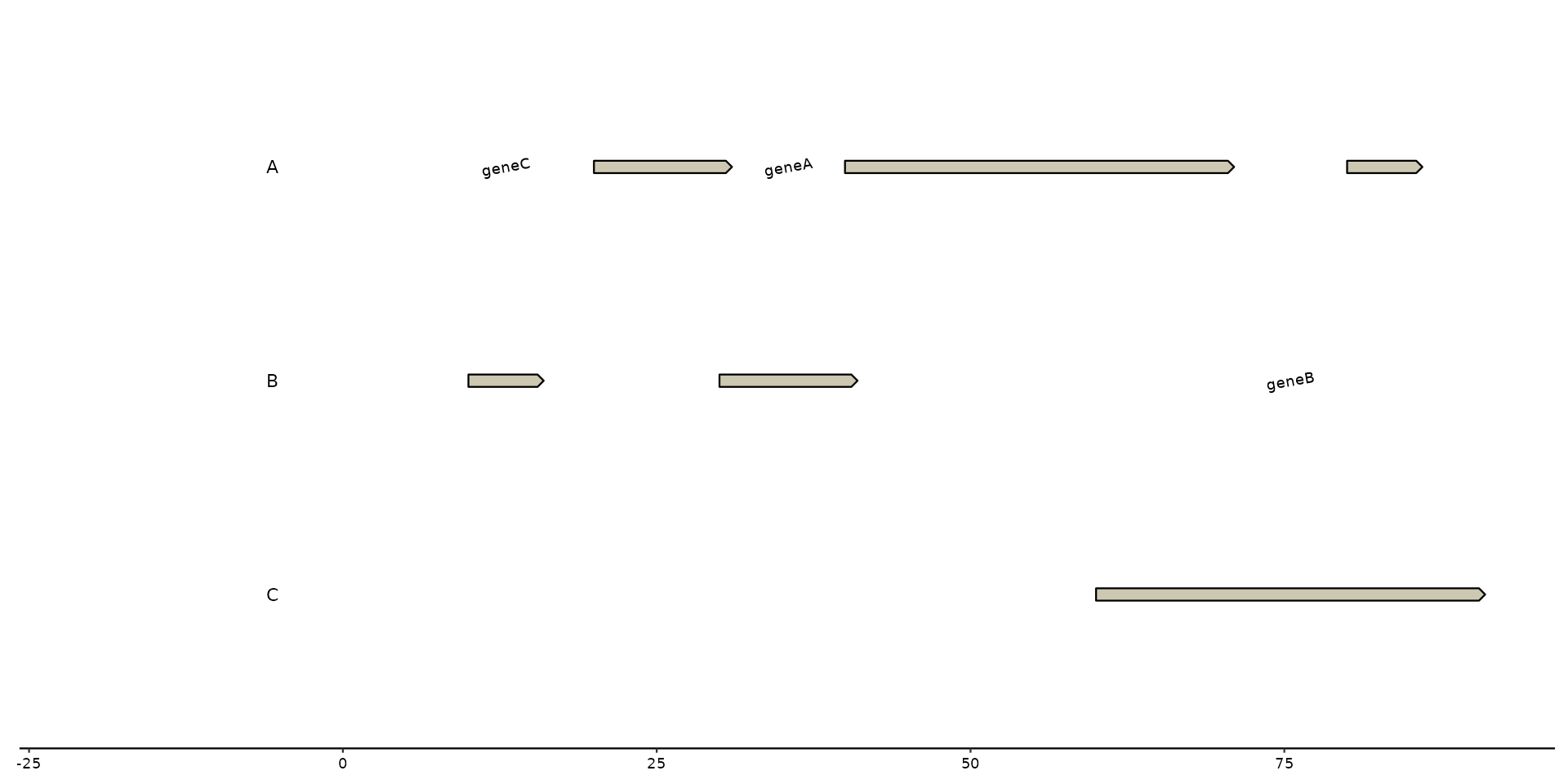 # labeling with manual input
plot + geom_gene_text(label = c("This", "is", "an", "example", "test", "test"))
# labeling with manual input
plot + geom_gene_text(label = c("This", "is", "an", "example", "test", "test"))